To the everyday animal lover, acorn worms have few things going for them. They are pretty hideous as a rule, and they don’t really do anything interesting. (If you count “eating mud all day” as interesting, you are probably a sediment ecologist…)
True to their mud-eating selves, though, acorn worms are great at muddying the water in the evo-devo world. You see, there’s this neat idea that goes back to the early 19th century called dorsoventral inversion. French naturalist Étienne Geoffroy Saint-Hilaire once noted that an arthropod looks rather like an upside down vertebrate in many ways. This diagram from the Wikipedia entry illustrates the concept rather nicely:

The pink and snot-green triangles on the side represent the expression levels of the key genes that determine where the back and belly sides go. dpp (decapentaplegic) is the fly homologue of BMP4 (or BMP2, 4 and 7, to be precise), and sog (short gastrulation) is the fly version of chordin. So not only do major structures like the central nervous system develop on opposite sides of the body, the developmental genetics are also upside down. It seems like a no-brainer. (Well, chordates had to somehow move the mouth back to the belly side, but hey, no big deal ;))
Then those evil acorn worms come in and complicate things.
Acorn worms belong to the phylum Hemichordata. Hemichordates are most closely related to echinoderms like sea urchins. Together with chordates (vertebrates, sea squirts and lancelets), they make up the deuterostomes. Fruit flies and most worms that aren’t “acorn”, on the other hand, are protostomes.
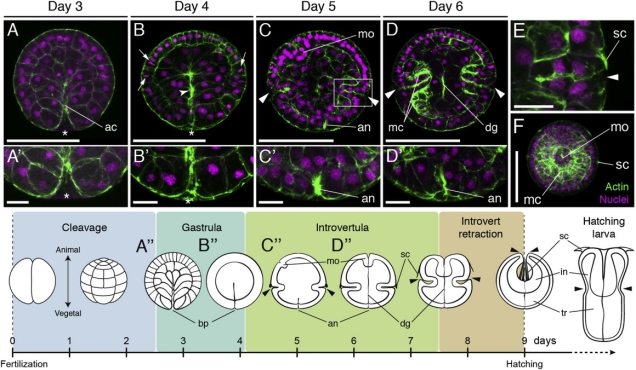
Whether acorn worms and other hemichordates have a central nervous system at all has been the subject of some debate. Recent evidence suggests that they do (Nomaksteinsky et al., 2009). They don’t even just have one, they have two of them, taking the form of nerve cords on the dorsal and ventral sides of the animal. The dorsal nerve cord forms in a way eerily similar to our own spinal cord. Chordates like ourselves undergo a process called neurulation, in which some of the ectoderm (“skin”) on the embryo’s back folds inwards to form a hollow tube that’s the precursor of the central nervous system. Well, look what happens in an acorn worm (figure from Luttrell et al., 2012):

The left side of this image depicts the process in the acorn worm Ptychodera flava (and shows you a young worm – innit cute?), while the right side illustrates what happens in a chick embryo. (I’m not sure why they didn’t use actual photos of chick neurulation, I don’t imagine they’d be very hard to come by…)
In chordates, the notochord (precursor of our spine) sends out chemical signals to direct nerve cord formation. Acorn worms don’t have anything notochord-like at the time the nerve cord develops, but somehow they make it look like neurulation anyway. If it looks like a duck (or chick, as the case may be) and quacks like one, it might just be one.
The problem?
It happens on the wrong side. Having read the two papers I’ve cited so far, my impression is that neurulation only happens with the dorsal cord. However, in other respects these animals share the arthropod, not the vertebrate, orientation. Most importantly, their genetic dorsoventral axis is oriented like that of arthropods and other protostomes, with BMP levels in the embryo highest on the dorsal side (Lowe et al., 2006). Chordate embryos can’t even make a nervous system at high BMP levels!
It seems that whatever happened to turn the ancestral bilaterian on its head, it wasn’t as simple as flipping the animal and relocating the mouth. The development of the central nervous system shares serious developmental genetic similarities among deuterostomes like us and protostomes like flies (Arendt and Nübler-Jung, 1999) or ragworms (Denes et al., 2007), indicating that if not a full-blown CNS, then at least the genetic pattern was present in our common ancestor.

The figure above is a schematic comparison of gene expression in the developing nervous systems of fruit flies (Drosophila), ragworms (Platynereis) and vertebrates from Denes et al. (2007). The diagram labelled Enteropneust illustrates the lack of data from acorn worms, which I don’t fully understand given that Lowe et al. (2006) actually studied several of the genes included in this figure. Speaking of that, acorn worms once again prove to be weird and confusing, since the genes in question aren’t expressed in anything like the longitudinal stripes seen in the other animals. In fact, Lowe et al. (2006) found that they aren’t obviously associated with either of the nerve cords.
To be precise, Lowe et al. worked under the assumption that acorn worms had no nerve cords, but if you look at their pictures the lack of resemblance is blindingly obvious. For example, Msx, the light grey gene on the Platynereis and vertebrate diagrams, goes all around acorn worm embryos in a relatively narrow ring. Nk2.2, the red gene, isn’t even expressed in the ectoderm (the embryonic “skin”), whereas central nervous systems invariably come from there. Did Lowe et al. get the wrong genes or what? Don’t think so, Msxes at least are pretty easy to recognise…
To summarise: acorn worm central nervous systems develop much like ours, but on the wrong side of the body, with none of the genetic similarities we share with animals much less closely related to us than acorn worms. To top that, the damn worms have another nerve cord on the opposite side, which doesn’t develop by neurulation unless I’ve misunderstood something.
I… have no idea what’s going on here. Damn you, nature, you need to work on your clarity.
***
References:
Arendt D & Nübler-Jung K (1999) Comparison of early nerve cord development in insects and vertebrates. Development 126:2309-2325
Denes AS et al. (2007) Molecular architecture of annelid nerve cord supports common origin of nervous system centralization in Bilateria. Cell 129:277-288
Lowe CJ et al. (2006) Dorsoventral patterning in hemichordates: insights into early chordate evolution. PLoS Biology 4:e291
Luttrell S et al. (2012) Ptychoderid hemichordate neurulation without a notochord. Integrative and Comparative Biology 52:829-834
Nomaksteinsky M et al. (2009) Centrarlization of the deuterostome nervous system predates chordates. Current Biology 19:1264-1269