I was planning to do more bioinformaticky stuff tonight, but then I saw Shimozono et al. (2013), and… SHINY!
I derive a particular joy from seeing neat methods, and what these guys did is pretty damn neat. They used genetic engineering and a clever trick with fluorescence to (almost) directly study an important but rather elusive molecule in vertebrate development.
Retinoic acid (RA) is related to vitamin A; in fact, it is synthesised from vitamin A by enzymes in our cells. It is what developmental biologists call a morphogen: a molecule that spreads through an embryo by diffusion, and influences development depending on its concentration. Among other things, RA is thought to be responsible for the subdivision of the embryonic body axis by Hox genes, and also the correct formation of somites, the basic repeating units that eventually form our spine.
So RA is pretty darn important, but it’s also a bit difficult to investigate. It’s a relatively small and simple molecule that isn’t encoded in the genome, so some of the popular tools for detecting important molecules don’t work on it. Its activity can be monitored indirectly, though. Retinoic acid works by binding to proteins called retinoic acid receptors (RARs), which then latch onto certain DNA sequences that regulate nearby genes. So you can, for example, construct a piece of DNA that responds to an activated RAR by producing a fluorescent protein. You can also examine the distribution of the enzymes that make and break down RA, the assumption being that this corresponds to the distribution of RA itself.
The Japanese team, however, created a modification of retinoic acid receptors that is basically a direct indicator of RA level. Their RARs have been engineered to glow in different ways depending on whether or not RA is bound to them. They were able to zap these miniature RA detectors into zebrafish embryos without affecting the little creatures’ development, creating a gentle way to monitor RA levels in live animals.
They exploited a fascinating phenomenon called fluorescence resonance energy transfer (FRET for short). FRET needs two fluorescent molecules that glow at different wavelengths, such that the wavelength one of them emits is the same that turns the other on. (Wikipedia tells me FRET is actually based on spooky quantum effects involving virtual particles rather than ordinary light travelling from molecule to molecule. Wow, I didn’t know that!)
If the two molecules are very, very close, the emissions of the first one can give the other enough energy to light up. You can detect this by shining the colour of light needed to excite the first molecule on your sample, but then also measuring the fluorescence from the second molecule. The ratio of Molecule 1 to Molecule 2 glow can tell you how much FRETting is going on.

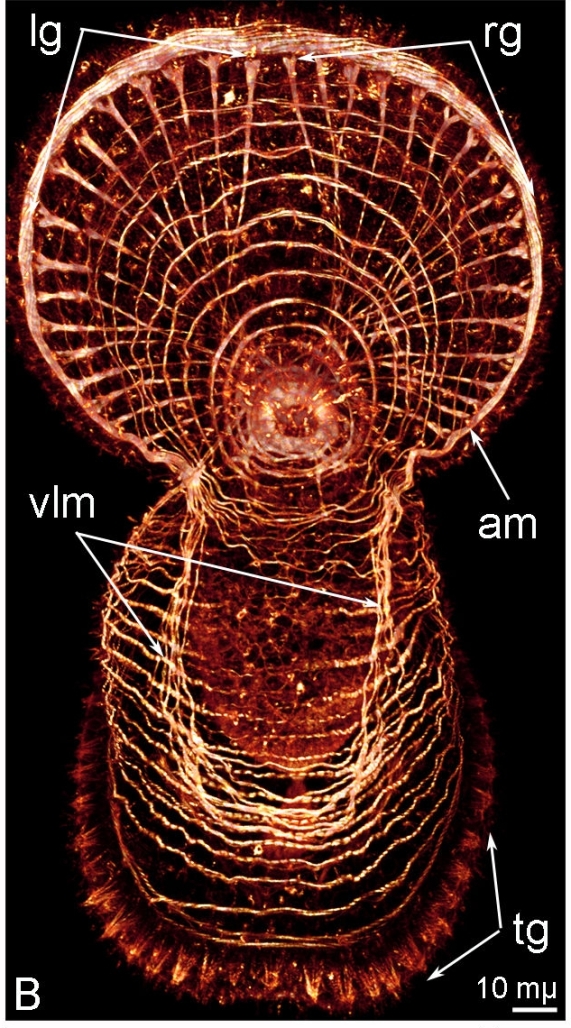
What Shimozono et al. did was to add the code for a FRET-capable pair of fluorescent proteins to various RAR genes. RARs change shape when retinoic acid binds to them, and in these engineered versions this means that they bring their fluorescent tags close enough for FRET to work. (The above figure, from Carr and Hetherington [2000], illustrates the principle – just substitute “Ca2+” with “RA”.) The scientists calibrated their little RA detectors by measuring how much FRET happened at various controlled RA concentrations first; this allowed them to turn FRET intensities into accurate measurements of RA. They then tested whether the detectors were truly RA-specific (and not activated by, say, vitamin A) by using them in fish embryos with their RA-making enzymes crippled.
Of course, they also got round to looking at the behaviour of RA during development, which was, after all, the point of their new toys. They did a basic visualisation of RA concentration throughout developing embryos – and confirmed that the established method of looking for the enzymes involved in RA synthesis and degradation is actually a decent substitute for measuring RA itself.
They then interfered with the production of a protein called FGF8 that is thought to regulate RA synthesis, and found that this altered the RA gradient – as well as the expression of the main enzyme that produces RA. Basically – the technique seems to work, and what it shows agrees with what we’ve thought about RA signalling. Hooray!
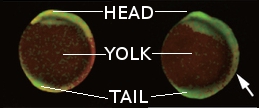
And, of course, they got pretty pictures like the ones below, coloured according to the amount of FRET (red = high, green = low) they measured. These two compare a normal embryo (left) and an embryo of the same age whose fgf8 gene has been messed with (right). If you have normal colour vision*, it’s pretty clear how the control embryo has this massive band of redness halfway down its body, and how even nearer the head and tail ends it’s more yellow than the sad green of the treated baby.
(I spliced these together from panels in an overwhelmingly massive figure and labelled them for those of you who don’t look at fish embryos much. No copyright infringement and no financial gain is intended, of course ;))
I think this whole thing is waaaaay cool. I wish I could come up with something clever like that. Oh well, at least I get to work with fluorescent things and take pretty glowy pictures every now and then. When I’m not neck deep in protein sequences and ‘omics data. 🙂
***
*Being a red-green colour blind developmental biologist must be a hell of a lot of fun. I just realised that pretty much everything involving fluorescence in biology is red, green or both – and developmental biologists love sticking fluorescent tags on everything. By the way, this particular figure could have been presented in any old combination of colours – they’re illustrating abstract numbers, not the actual colours of the specimens, which in this case would have been glowing in cyan and yellow. Of course, there’s probably a colour vision deficiency for every combination you can think of, so, uh. I probably overthunk this?
***
References:
Carr K & Hetherington A (2000) Calcium dynamics in single plant cells. Genome Biology 1:reports024
Shimozono S et al. (2013) Visualization of an endogenous retinoic acid gradient across embryonic development. Nature 496:363-366