… or wintertime holiday of your choice! Instead of a Christmas tree, I decorated one of Nick Hobgood’s gorgeous Christmas tree worms for you 😉

… or wintertime holiday of your choice! Instead of a Christmas tree, I decorated one of Nick Hobgood’s gorgeous Christmas tree worms for you 😉

If you are a bacterium, life can be pretty dangerous. Wherever you live, you are surrounded by millions of viruses that can inject you with their DNA and turn you into a helpless factory of these (bacteriophage T4 by Adenosine, Wikipedia):

(They may kill you, but at least they look badass…)
In response to the constant threat of deadly viruses, bacteria and archaea have evolved an ingenious defence mechanism called the CRISPR system. The CRISPR region in the genome consists of repetitions of a short sequence (the actual CRISPRs), alternating with spacers containing foreign (often viral) DNA the bacterium snatched from invaders. Spacers can be pulled out later and used to recognise and destroy the same invader.
If you are a CRISPR-enabled cell and survive a virus attack long enough to add the attacker to your “library”, you and your offspring are protected against the same virus forever. (OK, in reality, if a virus doesn’t show up for many generations, the microbes can afford to lose their “memory” of it to mutations. But provided that the virus is around and exerting a selection pressure, the immunity remains.)
The CRISPR system is fascinating for more than one reason. First, it’s essentially an adaptive immune system in some of the simplest organisms alive today. Adaptive immune systems are uncommon even in multicellular creatures. They are known from two groups of vertebrates (jawless and jawed vertebrates probably evolved them independently) and possibly brown algae (Zambounis et al., 2012).*
Second, it’s eerily “Lamarckian”. Microbes with CRISPR can, in a way, “direct” their own evolution. They are acquiring new adaptations over their lifetimes, and passing these on to their descendants. And they aren’t doing it in the way most inheritance of acquired traits works – instead of tagging their DNA with signals that remain highly flexible in the long term, they are actually permanently incorporating the new information into their genomes.
Parasites are notorious for evolving a way around anything a host can throw at them, though, and bacteriophages are no exception. A new study not-quite-published in Nature (Bondy-Denomy et al., 2012) reports viral genes that enable the viruses to break through CRISPR-based defences. They experimented on cultures of a strain of the bacterium Pseudomonas aeruginosa. Their bacteria went into the experiment infected with a variety of viruses, but the viruses were kept in their dormant form that doesn’t hurt the host cell. When these bacteria were exposed to active viruses, most of them could defend themselves just fine, while genetically engineered bacteria who lack a CRISPR system die like flies from the exact same pathogens.
However, a few of the bacteria were unable to resist the assault despite a fully intact immune system. Since the only difference between the different bacterial samples was the kind of dormant virus they hosted, the reason had to be something related to the viruses. A look at the viral genomes turned up several genes that almost literally held smoking guns. When added to CRISPR-sensitive viruses, they enabled them to kill bacteria they couldn’t otherwise harm. When deleted from their source viruses, they prevented the viruses from killing CRISPR-enabled but not CRISPR-disabled bacteria. A series of such experiments demonstrated that these “anti-CRISPR” genes helped viruses evade the immune systems of their hosts. Interestingly, they didn’t work against the closely related CRISPR system of E. coli – the cheat codes, so to speak, were highly specific to the game.
A really interesting thing about the CRISPR resistance genes is their possible origin. It seems Pseudomonas may have wrought its own doom in this case! When the researchers searched for sequences similar to the resistance genes, some of the sequences they caught were actually from other strains of Pseudomonas itself. A possible explanation is this: since CRISPR-based immunity can work on any foreign DNA, the original benefit of some anti-CRISPR genes may have been to prevent the destruction of bacterial genes after being passed to other bacteria. Then the viruses somehow got their hands on the sequences, hacking poor microbes with their own code.
Evolutionary arms races are such weird and crazy and fascinating things!
***
*I can’t really figure out whether some RNA interference based viral defence in plants counts. I’d have to go fact-hunting to find out if any of the interfering RNAs originate in a similar way to CRISPR spacers. The whole adaptive immunity angle is a digression, though, so I’m lazy enough to leave that up in the air.
***
References:
Bondy-Denomy J et al. (2012) Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature advance online publication, available 16/12/2012, doi: 10.1038/nature11723
Zambounis A et al. (2012) Highly dynamic exon shuffling in candidate pathogen receptors… what if brown algae were capable of adaptive immunity? Molecular Biology and Evolution 29:1263-1276
Gosh, can someone tell me if this is bullshit or if he has a point? O.o
It’s rather annoying when a paper comes out that basically threatens to turn what you think you know on its head, and you’re simply not equipped to evaluate its claims. This is the case with Retallack (2012). I’m fascinated by early animals, and endlessly bewildered by the strange fossils of the late Precambrian. While I’m aware that Ediacaran fossils have been interpreted as everything from microbial mats through animals to giant protists, I had the impression that the non-animal interpretations of iconic fossils like Dickinsonia, Spriggina, Parvancorina or Charniodiscus have slowly retreated to the fringe in the decades since their discovery.
And now this guy, whose name I’ve heard enough times to pay attention, gets into Nature arguing that the namesake formation of the Ediacaran period actually originated on dry land, and the iconic fossils are preserved in a manner more like plants, fungi or lichens than animals.
The paltry one semester of introductory geoscience I did years ago is nowhere near enough to comment on all the stuff he says about soils and microbial mats and preservation. I feel completely out of my depth, rocking precariously at the mercy of the waves…
Obviously, this assessment of the original Ediacara site doesn’t affect every fossil site from the period. The latest Precambrian reefs of the Nama Group remain marine reefs containing the remains of unknown animals that grew some of the first mineralised skeletons.
My big question at the moment is how Retallack would interpret the preservation of the White Sea assemblage. This contains similar kinds of fossils to the sites he’s reinterpreted as terrestrial. There’s Dickinsonia and several others like it, there’s Parvancorina, there’s Cyclomedusa*. And this is where hundreds of specimens of my Platonic love Kimberella come from, often associated with crawling and feeding traces. That guy moved around and grazed – plants and lichens seldom do such things! So was Kimberella a land animal? That would be the biggest palaeontological sensation of the decade if not the century. Or did dickinsoniids etc. occur both on land and underwater? Or did the White Sea fossils span a wide variety of environments? (I’m not sure about the distribution of the various White Sea fossils relative to each other…)
Oh my. I wonder what will come out of this. Publication in Nature makes it dead certain that any expert who’d vehemently disagree will find the article. Let’s pull out the pop corn and watch…
***
*It’s slightly odd that he seemingly treats Cyclomedusa and other “medusoid” fossils as though most people considered them jellyfish. That may have been their original interpretation, but I thought it was widely discredited now.
***
Reference:Retallack GJ (2012) Ediacaran life on land. Nature advance online publication available 12/12/12, doi:10.1038/nature11777
My heart is a bit broken.
On my way to work this morning, I saw a crow tearing into the corpse of a small bird, possibly a greenfinch from the brief look I got. I don’t know if the crow had killed it or if it had dropped dead all by itself, but in the end it doesn’t really matter.
I love crows and I know they are opportunistic bastards that will eat anything, but I’ve never actually seen one feast on a small bird. And small birds are my little feathered antidepressants. It’s like seeing your beloved cat dismember your pet hamster.
I’ll console myself with Andreas Trepte’s gorgeous photos of a non-dismembered greenfinch. Isn’t he a beauty!
I went to a talk by Paul Brakefield today. I loved it. It was a summary of decades of work on butterfly eyespots covering just about everything about them. There was development and evolvability, ecology and adaptation, courtship and speciation, really all an evolutionary biologist could wish for. Yay!
Nonetheless, my biggest squee moment in the entire talk was the first slide, which contained a picture of Kjell Sandved’s butterfly alphabet.
Back in the sixties, this guy who had taken like one photo in his entire life realised he could spell things out with patterns on butterflies’ wings. So he went and learned photography and travelled the world to hunt down the entire Latin alphabet plus symbols and Arabic numerals. All on little scaly lepidopteran wings. I have no words for how fucking cool that is. I don’t want to post the alphabet itself since it’s copyrighted, but do a google search or go here and you’ll see. You’ll see.
Wow.
Damn. Mistaking evolution for a ladder with us on top is something I fully expect from people who don’t study it for a living, but when evolutionary scientists make that mistake, it drives me apeshit. And they do it all the fucking time.
I don’t think most of them are aware of it. You’ve got to be really watching for the trap to have a chance of avoiding it. I slip every now and then, and then I spot it and rage at myself and get deeply philosophical about human nature and such. It’s such an easy and convenient thing to do. (Think of evolution as a ladder, not get philosophical, I mean.) It’s the way we’ve been conditioned to think since the first time we heard about evolution.
For most of the history of biology, no one blinked twice if you talked with culturally sanctioned anthropocentrism about “lower animals” or “higher vertebrates”. Evolution was a highway of progress, and some creatures just got further along than others. Naturally, we were speeding along right at the front.
Nowadays, I think most biologists who have to consider evolution in their work would tell you that evolution doesn’t work like that. The papers I read rarely contain such explicit references to the “march of progress”. (Can I call it the MOP?) However, that doesn’t mean the references are gone. They’ve just become so subtle that, I suspect, not even the people who make them realise they’re there.
It’s “basal lineages”. “Phylogenetically more primitive” creatures. Or “early-branching organisms”. Or “evolutionary old animals”. All of these are real terms used in real papers published this year. They aren’t restricted to bad papers. And if you stop to think about it, none of them make any goddamned sense.
Let’s picture an evolutionary tree first. I can’t really use my usual tree with all its question marks, but the one below, which I nicked from Srivastava et al. (2008), will do:
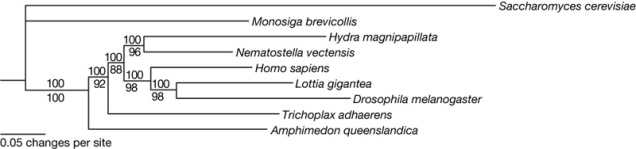
(The species from top to bottom are: brewer’s yeast, a choanoflagellate, this tentacled little guy, a sea anemone, humans, a limpet, everyone’s favourite fruit fly, the Blob, and a sponge.)
The “base” of the tree is to the left, where animals, Monosiga and fungi have their last common ancestor. (That was a long time ago.) “Basal” means close to the base. The branching point (node) that separates animals from the non-animals at the top is the basalmost node in this tree. The node that separates the sponge from the other animals is also a pretty basal node. The creature that gave rise to both sponges and other animals was a truly basal animal.
Now, which is the basal lineage?
The correct answer is “relative to what?”
Every node divides the tree into two lineages. It doesn’t make any sense to say that one of them is more basal than the other. There’s a basal node in the tree of animals. Sponges are on one side of that, the rest of the animals are on the other. If you take a vertebrate species, sponges are the last animal lineage you’ll encounter if you trace its ancestry back towards the base of the tree. If you take a sponge species, the lineage with vertebrates (and lots of other things) on it will be the last.
“Basal lineage” depends on your point of view.
Maybe actually taking the sponge point of view will help illustrate this. This tree comes from a paper about sponges (Sperling et al., 2010):
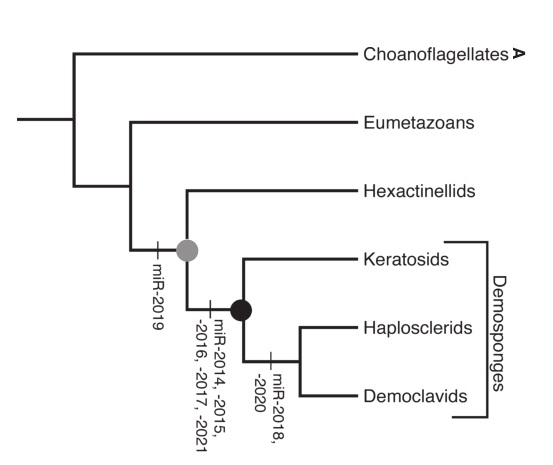
Unlike the previous tree, its branches are labelled with larger groups rather than species, but these represent more or less the same range of creatures. Monosiga from tree one is a choanoflagellate. Amphimedon is a haplosclerid demosponge, on the second branch from the bottom. Every other animal from the first tree is compressed down into that one branch labelled “Eumetazoans”. (OK, Trichoplax is not a eumetazoan, but that’s a technicality that doesn’t affect the point.) From this angle, it’s rather harder to see sponges as a basal animal lineage!
Equally, sponges are just as old as non-sponge animals, so calling them “old” is a tad dodgy. Here, you could argue that sponges have been around longer than, say, vertebrates, which is true to the best of our knowledge. In that sense, “sponges” is an older lineage than “vertebrates”. But that only means that “sponges” should be compared to “non-sponges” rather than “vertebrates”, and anyone making such comparisons should be as aware of the diversity lurking within sponges as they are of the diversity of other animals.
The “evolutionary old animals” quote actually comes from a paper that looked at stem cell genes in Hydra to understand the evolution of stem cells in animals. (Hemmrich et al., 2012). It’s not comparing cnidarians (the phylum hydras belong to) to something genuinely younger than them. I can’t resist quoting the whole offending sentenc:
Our observations provided new and comprehensive insight into the complex network that orchestrates patterning and tissue homeostasis in an evolutionary old animal that branched off almost 600 million years ago. (p3277)
Honestly, what does that even mean? Branched off from what?
OK, I know it means from our own ancestors. But my point is that this should not be taken for granted, and if you do take a human-centric point of view, you should bloody well make that explicit. You should not write as though evolution had some sort of “main branch” leading to us from which things split every now and then. Lineages split from each other.
You might think that I’m being pedantic just to have an excuse to rant, but the implicit views underlying examples like the above have real consequences for the study of evolution. Namely, they might lead scientists to assume that representatives of “basal” lineages got stuck in the Precambrian and could just stand in for their distant ancestors. This is dangerous.
Take sponges. Yes, in many respects they probably resemble the first animals more than we do. Chances are those ancient animals didn’t have sophisticated organs and like two hundred different cell types. However, chances also are that they were made of distinct cells rather than huge merged syncytia, and that they didn’t have elaborate skeletons made of some sort of mineral, both of which are properties of many sponges. All animals alive today had exactly the same amount of time to evolve their own quirks since their last common ancestor. We shouldn’t just assume that anything “simple” in an animal we regard as “basal” is inherited straight from that ancestor just because it fits our favourite story.
Case in point: the Amphimedon genome was found to be impoverished in many families of developmentally important “master” genes, and this fit nicely into the prevailing view of the increasing complexity of animals throughout their history (Larroux et al., 2008). But it’s likely that at least some of those genes were actually lost by Amphimedon‘s ancestors and not gained by ours (Mendivil Ramos et al., 2012). Assuming that “basal” (relative to us) means “similar to ancestor X” can very easily lead to unwarranted conclusions, and that can hinder our ability to figure out what really happened. To me, that’s a big deal.
***
References:
Hemmrich G et al. (2012) Molecular signatures of the three stem cell lineages in Hydra and the emergence of stem cell function at the base of multicellularity. Molecular Biology and Evolution 29:3267-3280
Larroux C et al. (2008) Genesis and expansion of metazoan transcription factor gene classes. Molecular Biology and Evolution 25:980-996
Mendivil Ramos O et al. (2012) Ghost loci imply Hox and ParaHox existence in the last common ancestor of animals. Current Biology 22:1951-1956
Sperling EA et al. (2010) Where’s the glass? Biomarkers, molecular clocks, and microRNAs suggest a 200-Myr missing Precambrian fossil record of siliceous sponge spicules. Geobiology 8:24-36
Srivastava M et al. (2008) The Trichoplax genome and the nature of placozoans. Nature 454:955-960