ParaHox genes are a bit like the underappreciated sidekicks of Hox genes. Or little sisters, as the case may be, since the two families are closely related. Hox genes are probably as famous as anything in evo-devo. Being among the first genes controlling embryonic development to be (a) discovered, (b) found to be conserved between very distantly related animals, they are symbolic of the late 20th century evo-devo revolution.
ParaHoxes get much less attention despite sharing some of the most exciting properties of Hox genes. Like those, they are involved in anteroposterior patterning – that is, partitioning an embryo along its head to tail axis. Also like Hox genes, they are often neatly clustered in the genome, and when they are, they tend to be expressed in the same order (both in space and time) in which they sit in the cluster*. Their main ancestral roles for bilaterian animals seem to be in patterning the gut and the central nervous system (Garstang and Ferrier, 2013).
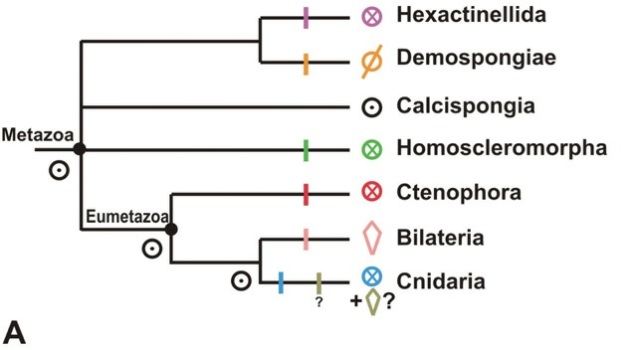
There are three known types of ParaHox gene, which are generally thought to be homologous to specific Hox subsets of Hox genes – by the most accepted scheme, Gsx is the closest sister of Hox1 and Hox2, Xlox is closest to Hox3, and Cdx to Hox9 and above. It is abundantly clear that Hoxes and ParaHoxes are closely related, but there has been a bit of debate concerning the number of genes in the ancestral gene cluster that gave rise to both – usually called “ProtoHox” (Garcia-Fernàndez, 2005).
Another big question about these genes is precisely when they originated, and in this regard, ParaHox genes are proving much more interesting than Hoxes. You see, there are plenty of animals with both Hox and ParaHox genes, which is what you’d expect given the ProtoHox hypothesis, but there are also animals with only ParaHoxes. If there really was a ProtoHox gene/cluster that then duplicated to give rise to Hoxes and ParaHoxes, then lone ParaHoxes (or Hoxes for that matter) shouldn’t happen – unless the other cluster was lost along the way.
So a suspiciously Gsx-like gene in the weird little blob-creature Trichoplax, which has nothing remotely resembling a Hox gene, was a big clue that (a) Hox/ParaHox genes might go back further in animal evolution than we thought, (b) the loss of the entire Hox or ParaHox cluster is totally possible**, despite how fundamental these genes appear to be for correctly building an animal.
I wrote (at length) about a study by Mendivil Ramos et al. (2012), which revealed that while Trichoplax had no Hox genes and only one of the three types of ParaHox gene, it preserved the more or less intact genomic neighbourhoods in which Hox and ParaHox clusters are normally situated. One of the more interesting results of that paper was that the one sponge genome available at the time – that of Amphimedon queenslandica, which had no trace of either Hox or ParaHox genes – also contained statistically significant groupings of Hox and ParaHox neighbour genes, as if it had a Hox neighbourhood and a ParaHox neighbourhood, but the Hoxes and ParaHoxes themselves had moved out.
That study thus pointed towards an intriguing hypothesis, previously championed by Peterson and Sperling (2007) based solely on gene phylogenies: sponges once did have Hox and ParaHox genes/clusters, which at least some of them later lost. This would essentially mean that the two gene clusters go straight back to the origin of animals if not further***, and we may never find any surviving remnant of the ancestral ProtoHox cluster, since the closest living relatives of animals have neither the genes nor their neighbourhoods (that we know of).
Hypotheses are nice, but as we know, they do have a tendency to be tragically slain by ugly facts. Can we further test this particular hypothesis about sponges? Are there facts that could say yay or nay? (Of course there are. I wouldn’t be writing this otherwise 😉 )
I keep saying that we should always be careful when generalising from one or a few model organisms, that we ignore diversity in the animal kingdom at our own peril, and that “distantly related to us” = “looks like our distant ancestors” is an extremely dodgy assumption. Well, here’s another lesson in that general vein: unlike Amphimedon, some sponges have not just the ghosts of vanished ParaHox clusters, but intact, honest to god ParaHox genes!
It’s calcareous sponges again. Sycon ciliatum and Leucosolenia complicata, two charming little calcisponges, recently had their genomes sequenced (alas, they weren’t yet public last time I checked), and since then, there’s been a steady stream of “cool stuff we found in calcisponge genomes” papers from Maja Adamska’s lab and their collaborators. I’ve discussed one of them (Robinson et al., 2013), in which the sponges revealed their rather unhelpful microRNAs, and back in October (when I was slowly self-destructing from thesis stress), another study announced a couple of delicious ParaHoxes (Fortunato et al., 2014).
(Exciting as it is, the paper starts by tickling my pet peeves right off the bat by calling sponges “strong candidates for being the earliest extant lineage(s) of animals”… I suppose nothing can be perfect… *sigh*)
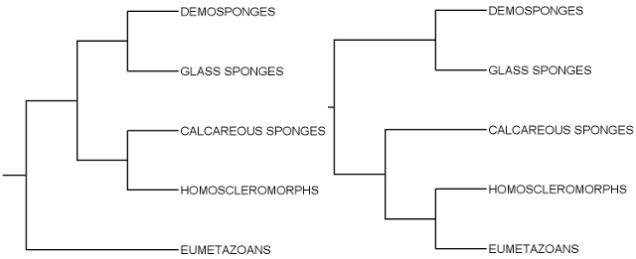
The study actually covers more than just (Para)Hox genes; it looks at an entire gene class called Antennapedia (ANTP), which includes Hoxes and ParaHoxes plus a handful of related families I’m far less interested in. Sycon and Leucosolenia don’t have a lot of ANTP genes – only ten in the former and twelve in the latter, whereas a typical bilaterian like a fruit fly or a lancelet has several times that number – but from phylogenetic analyses, these appear to be a slightly different assortment of genes from those present in Amphimedon, the owner of the first sequenced sponge genome. This picture is most consistent with a scenario in which all of the ANTP genes in question were present in our common ancestor with sponges, and each sponge lineage lost some of them independently. (You may not realise this until you start delving into the history of various gene families, but genes come and go a LOT in evolution.)
Sadly, many of the branches on these gene trees are quite wonky, including the one linking a gene from each calcisponge to the ParaHox gene Cdx. However, somewhat fuzzy trees are not the only evidence the study presents. First, the putative sponge Cdxes possess a little motif in their protein sequences that is only present in a handful of gene families within the ANTP class. If you take only these families rather than everything ANTP and make trees with them, the two genes come out as Cdx in every single tree, and with more statistical support than the global ANTP trees gave them. Another motif they share with all Hoxes, ParaHoxes and a few of their closest relatives, but not with other ANTP class families.
Second, at least the gene in Sycon appears to have the right neighbours (Leucosolenia was not analysed for this). Since the Sycon genome sequence is currently in pieces much smaller than whole chromosomes, only four or so of the genes flanking ParaHox clusters in other animals are clearly linked to the putative Cdx in the sponge. However, when the researchers did the same sort of simulation Mendivil Ramos et al. (2012) did for Amphimedon, testing whether Hox neighbours and ParaHox neighbours found across all fragments of the genome are (a) close to other Hox/ParaHox neighbours or randomly scattered (b) mixed or segregated, they once again found cliques of genes with little overlap, indicating the once-existence of separate Hox and ParaHox clusters.
Fortunato et al. (2014) also examined the expression of their newfound Cdx gene, and found it no less intriguing than its sequence or location in the genome, although their description in the paper is very limited (no doubt because they’re trying to cram results on ten genes into a four-page Nature paper). The really interesting activity they mention and picture is in the inner cell mass of the young sponge in its post-larval stages – the bit that develops into the lining of its feeding chambers. Which, Adamska’s team contend, may well be homologous to our gut lining. In bilaterians, developing guts are one of the major domains of Cdx and ParaHox genes in general!
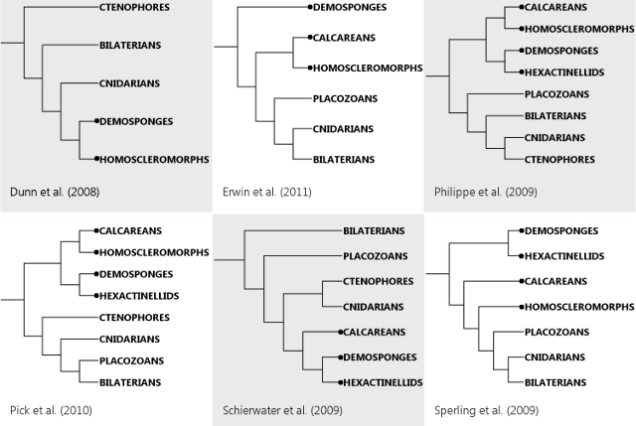
So at least three different lines of evidence – sequence, neighbours and expression – make this picture hang together quite prettily. It’s incredibly cool – the turning on their heads of long-held assumptions is definitely the most exciting part of science, I say! On the other hand, it’s also a little disheartening, because now that everyone in the animal kingdom except ctenophores has definitive ParaHox genes and at least the empty seats once occupied by Hox genes, are we ever going to find a ProtoHox thingy? May it be that it’ll turn up in one of the single-celled beasties people like Iñaki Ruiz-Trillo are sequencing? That would be cool and weird.
The coolest twist on this story, though, would be to discover traces of ProtoHoxes in a ctenophore, since solid evidence for ProtoHox-wielding ctenophores would (a) confirm the strange and frankly quite dubious-sounding idea that ctenophores, not sponges, are the animal lineage farthest removed from ourselves, (b) SHOW US A FREAKING PROTOHOX CLUSTER. (*bounces* >_> Umm, * cough* OK, maturity can suck it 😀 ) However, given how horribly scrambled at least one ctenophore genome is (Ryan et al., 2013), that’s probably a bit too much to ask…
***
Notes
*Weirdly, the order of expression in time is the opposite of that of the Hox cluster. In both clusters, the “anterior” gene(s), i.e. Hox1-2 or Gsx, are active nearest the front of the embryo, but while anterior Hox genes are also the earliest to turn on, in the ParaHox cluster the posterior gene (Cdx) wakes up first. /end random trivia
**Of course we’ve long known that losing a Hox cluster is not that big a deal, but previously, all confirmed losses occurred in animals with more than one Hox cluster to begin with – a fish has plenty of Hox genes left even after chucking an entire set of them.
***With the obligatory ctenophore caveat…
***
References
Fortunato SAV et al. (2014) Calcisponges have a ParaHox gene and dynamic expression of dispersed NK homeobox genes. Nature 514:620-623
Garcia-Fernàndez J (2005) The genesis and evolution of homeobox gene clusters. Nature Reviews Genetics 6:881-892
Garstang M & Ferrier DEK (2013) Time is of the essence for ParaHox homeobox gene clustering. BMC Biology 11:72
Mendivil Ramos O et al. (2012) Ghost loci imply Hox and ParaHox existence in the last common ancestor of animals. Current Biology 22:1951-1956
Peterson KJ & Sperling EA (2007) Poriferan ANTP genes: primitively simple or secondarily reduced? Evolution and Development 9:405-408
Robinson JM et al. (2013) The identification of microRNAs in calcisponges: independent evolution of microRNAs in basal metazoans. Journal of Experimental Zoology B 320:84-93
Ryan JF et al. (2013) The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 342:1242592