The Mammal has emerged from a thesis-induced supermassive black hole and a Christmas-induced food coma, only to find that in the month or so that she spent barely functional and buried in chapters covered in the supervisor’s dreaded Red Pen, things actually happened in the world outside. This, naturally, manifested in thousands of items feeling thoroughly neglected in RSS readers and email inboxes. (Jesus. How many times have I vowed never to neglect my RSS feed again? Oh well, it’s not like unemployment is such a busy occupation that I can’t deal with a measly two and a half thousand articles 😛 )
… earlier tonight, the paragraph here said I wasn’t doing a proper post yet, “just pointing out” a couple of the cooler things I’ve missed. Then somehow this thing morphed into a 1000+ word post that goes way beyond “pointing things out”. It’s almost like I’ve been itching to write something that isn’t my thesis. >_>
So the first cool thing I wanted to “point out” is the genome paper of the centipede Strigamia maritima, which is a rather nondescript little beast hiding under rocks on the coasts of Northwest Europe. This is the first sequenced genome of a myriapod – the last great class of arthropods to remain untouched by the genome sequencing craze after many genomes from insects, crustaceans and chelicerates (spiders, mites and co.). The genome sequence itself has been available for years (yay!), but its “official” paper (Chipman et al., 2014) is just recently out.
Part of the appeal of Strigamia – and myriapods in general – is that they are considered evolutionarily conservative for an arthropod. In some respects, the genome analysis confirms this. Compared to its inferred common ancestor with us, Strigamia has lost fewer genes than insects, for example. Quite a lot of its genes are also linked together similarly to their equivalents in distantly related animals, indicating relatively little rearrangement in the last 600 million years or so. But this otherwise conservative genome also has at least one really unique feature.
Specifically, this centipede – which is blind – has not only lost every bit of DNA coding for known light-sensing proteins, but also all known genes specific to the circadian clock. In other animals, genes like clock and period mutually regulate one another in a way that makes the abundance of each gene product oscillate in a regular manner (this is about the simplest graphical representation I could find…). The clock runs on a roughly daily cycle all by itself, but it’s also connected to external light via the aforementioned light-sensing proteins, so we can constantly adjust our internal rhythms according to real day-night cycles.
There are many blind animals, and many that live underground or otherwise find day and night kind of irrelevant, but even these are often found to have a functioning circadian clock or keep some photoreceptor genes around. However, based on the genome data, our favourite centipede may be the first to have completely lost both. The authors of the genome paper hypothesise that this may be related to the length of evolutionary time the animals have spent without light. Things like mole rats are relatively recent “inventions”. However, the geophilomorph order of centipedes, to which Strigamia belongs, is quite old (its most likely sister group is known from the Carboniferous, so they’re probably at least that ancient). Living geophilomorphs are all blind, so chances are they’ve been that way for the last 300+ million years.
Nonetheless, the authors also note that geophilomorphs are still known to avoid light – the question now is how the hell they do it… And, of course, whether Strigamia has a clock is not known – only that it doesn’t have the clock we’re used to. We also have no idea at this point how old the gene losses actually are, since all the authors know is that one other centipede from a different group has perfectly good clock genes and opsins.
In comparison with fruit flies and other insects, the Strigamia genome also reveals some of the ways in which evolutionary cats can be skinned in multiple ways. There is an immune-related gene family we share with arthropods and other animals, called Dscam. The product of this gene is involved in pathogen recognition among other things, and in flies, Dscam genes are divided into roughly 100 chunks or exons, most of which are are found in clusters of variant copies. When the gene is transcribed, only one of these copies is used from each such cluster, so in practical terms the handful of fruit fly Dscam genes can encode tens of thousands of different proteins, enough to adapt to a lot of different pathogens.
A similar arrangement is seen in the closely related crustaceans, although with fewer potential alternative products. In other groups – the paper uses vertebrates, echinoderms, nematodes and molluscs for comparison – the Dscam family is pretty boring with at most one or two members and none of these duplicated exons and alternative splicing business. However, it looks like insects+crustaceans are not the only arthropods to come up with a lot of DSCAM proteins. Strigamia might also make lots of different ones (“only” hundreds in this case), but it achieved this by having dozens of copies of the whole gene instead of performing crazy editing feats on a small number of genes. Convergent evolution FTW!
Before I paraphrase the entire paper in my squeeful enthusiasm (no, seriously, I’ve not even mentioned the Hox genes, and the convergent evolution of chemoreceptors, and I think it’s best if I shut up now), let’s get to something else that I can’t not “point out” at length: a shiny new vetulicolian, and they say it’s related to sea squirts!
Vetulicolians really deserve a proper discussion, but in lieu of a spare week to read up on their messiness, for now, it’s enough to say that these early Cambrian animals have baffled palaeontologists since day one. Reconstructions of various types look like… a balloon with a fin? Inflated grubs without faces? I don’t know. Drawings below (Stanton F. Fink, Wikipedia) show an assortment of the beasts, plus Yunnanozoon, which may or may not have something to do with them. Here are some photos of their fossils, in case you wondered.
They’re certainly difficult creatures to make sense of. Since their discovery, they’ve been called both arthropods and chordates, and you can’t get much farther than that with bilaterian animals (they’re kind of like the Nectocaris of old, come to think of it…).
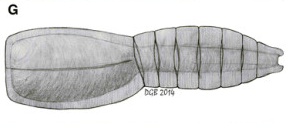
The latest one was dug up from the Emu Bay Shale of Australia, the same place that yielded our first good look at anomalocaridid eyes. Its newest treasure has been named Nesonektris aldridgei by its taxonomic parents (García-Bellido et al., 2014), and it looks something like this (Diego García-Bellido’s reconstruction from the paper):
In other words, pretty typical vetulicolian “life but not as we know it”, at first glance. Its main interest lies in the bit labelled “nc” in the specimens shown below (from the same figure):
This chunky structure in the animal’s… tail or whatever is a notochord, the authors contend. Now, only one kind of animal has a notochord: a chordate. (Suspicious annelid muscle bundles notwithstanding. Oh yeah, I also wanted to post on Lauri et al. 2014. Oops?) So if this thing in the middle of Nesonektris’s tail is a notochord, then at the very least it is more closely related to chordates than anything else.
Why do they think it is one? Well, there are several long paragraphs devoted to just that, so here goes a summary:
1. It’s probably not the gut. A gut would be the other obvious ID, but it doesn’t fit very well in this case. Structures interpreted as guts in other vetulicolians – which sometimes contain stuff that may be half-digested food – (a) start in the front half of the body, where the mouth is, (b) constrict and expand and coil and generally look much floppier than this, (c) don’t look segmented, (d) sometimes occur alongside these tail rod-like thingies, so probably aren’t the same structure.
2. It positively resembles modern half-decayed notochords. The notochords of living chordates are long stacks of (muscular or fluid-filled) discs, which fall apart into big blocks as the animal decomposes after death. Here’s what remains of the notochord of a lamprey after two months for comparison (from Sansom et al. (2013)):
This one isn’t as regular as the blockiness in the fossils, I think, but that could just be the vetulicolians not being quite as rotten.
There is, of course, a but(t). To be precise, there are also long paragraphs discussing why the structure might not be a notochord after all. It’s much thicker than anything currently interpreted as such in reasonably clear Cambrian chordates, for one thing. Moreover, it ends right where the animal does, in a little notch that looks like a good old-fashioned arsehole. By the way, the paper notes, vetulicolian tails in general don’t go beyond their anuses by any reasonable interpretation of the anus, and a tail behind the anus is kind of a defining feature of chordates, though this study cites a book from the 1970s claiming that sea squirt larvae have a vestigial bit of proto-gut going all the way to the tip of the tail. (I suspect that claim could use the application of some modern cell labelling techniques, but I’ve not actually seen the book…)
… and there is a phylogenetic analysis, in which, if you interpret vetulicolians as deuterostomes (which impacts how you score their various features), they come out specifically as squirt relatives whether or not you count the notochord. I’m never sure how much stock to put in a phylogenetic analysis based on a few bits of anatomy gleaned from highly contentious fossils, but at least we can say that there are other things – like a hefty cuticle – beyond that notochord-or-not linking vetulicolians to a specific group of chordates.
Having reached the end, I don’t feel like this paper solved anything. Nice fossils either way 🙂
And with that, I’m off. Maybe next time I’ll write something that manages to be about the same thing throughout. I’ve been thinking that I should try to do more posts about broader topics rather than one or two papers (like the ones I wrote about ocean acidification or homology versus developmental genetics), but I’ve yet to see whether I’ll have the willpower to handle the necessary reading. I’m remarkably lazy for someone who wants to know everything 😀
(Aside: holy crap, did I ALSO miss a fucking Nature paper about calcisponges’ honest to god ParaHox genes? Oh my god, oh my GOD!!! *sigh* This is also a piece of incredibly exciting information I’ve known for years, and I miss it when it actually comes out in a journal bloody everyone reads. You can tell I’ve been off-planet!)
References:
Chipman AD et al. (2014) The first myriapod genome sequence reveals conservative arthropod gene content and genome organisation in the centipede Strigamia maritima. PLoS Biology 12:e1002005
García-Bellido DC et al. (2014) A new vetulicolian from Australia and its bearing on the chordate affinities of an enigmatic Cambrian group. BMC Evolutionary Biology 14:214
Lauri A et al. (2014) Development of the annelid axochord: insights into notochord evolution. Science 345:1365-1368
Sansom RS et al. (2013) Atlas of vertebrate decay: a visual and taphonomic guide to fossil interpretation. Palaeontology 56:457-474