What the hell is up with lamprey Hox clusters?

Lampreys are among the few living jawless vertebrates, creatures that parted evolutionary ways with our ancestors somewhere on the order of 500 million years ago. If you want to know where things like jaws, paired fins or our badass adaptive immune systems came from, a vertebrate that doesn’t possess some of these things and may have diverged from the rest of the vertebrates soon after others originated is just what you need for comparison.
The vertebrate fossil record is pretty rich thanks to us having hard tissues, so a lot can be inferred about these things from the wealth of extinct fishes we have at our disposal. However, there are times when comparisons of living creatures are just as useful, if not more, than examinations of fossils. (Fossils, for example, tend not to have immune systems. ;))
One of the things you absolutely need a living animal to study is, of course, genome evolution. Vertebrates – well, at least jawed vertebrates – are now generally accepted to have the remnants of four genomes. Our long-gone ancestors underwent two rounds of whole genome duplication. Afterwards, most of the extra genes were lost, but evidence for the duplications can still be found in the structure of our genomes, where entire recognisable gene neighbourhoods of our close invertebrate relatives often still exist in up to four copies (Putnam et al., 2008).
Among these neighbourhoods are the four clusters of Hox genes most groups of jawed vertebrates possess. A “normal” animal like a snail or a centipede only has one of these. Since Hox genes are involved in the making of body plans, you have to wonder how suddenly having four sets of them and other developmental “master genes” might have influenced the evolution of vertebrate bodies.
Of course, to guess that, you need to know precisely when these duplications happened. That’s where lampreys come in: their lineage branched off from our definitely quadruple-genomed one after the next closest, definitely single-genomed group. But was it before, between, or after, the two rounds of duplication?
A few years ago, a phylogenetic analysis of 55 gene families by Kuraku et al. (2009) suggested that the lamprey-jawed vertebrate split happened after the 2R. Just this year, the genome of the sea lamprey Petromyzon marinus was finally published (Smith et al., 2013), and its authors agreed that yes, lampreys probably split off from us post-2R. (I don’t entirely get all the things they did to arrive at this conclusion. Groups of linked genes show up again, among other approaches.)
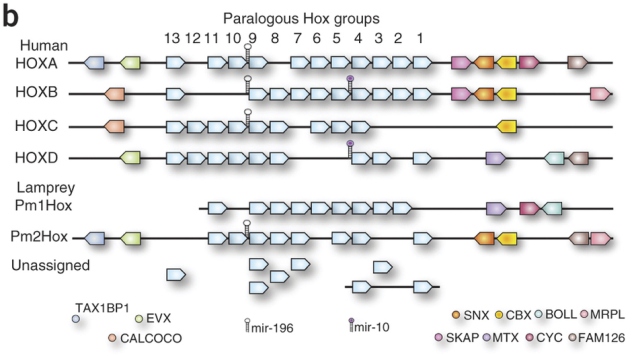
However, that isn’t the whole story, the latest lamprey genomics paper argues (Mehta et al., 2013). The P. marinus genome assembly couldn’t stitch all the Hox clusters properly together. There were two that sat on nice big scaffolds with the whole row of Hox genes and a few of their neighbours, and then there were a bunch of “loose” Hox genes that they couldn’t link to anything (diagram comparing humans and P. marinus below from Smith et al., 2013; the really pale blue boxes under the numbers represent Hox genes):
Given that Hox9 genes exist in four copies in this species, it seems like there may be four clusters. However, in hagfish, the other kind of living jawless vertebrate, a study found Hox genes that seemed to have as many as seven copies (Stadler et al., 2004). Another round of duplication? It wouldn’t be unheard of. Most teleosts, which include most of the things we call “fish” in everyday parlance, have seven Hox clusters courtesy of an extra genome duplication and loss of one cluster*. Salmon and kin have thirteen, after yet another duplication. Maybe hagfish also had another one – but did lampreys? How many more clusters do those lonely Hox genes belong to?
Mehta et al. hunted down the Hox clusters of Japanese lampreys (Lethenteron japonicum), hoping to pin down exactly how many there were. They used large chunks of DNA derived partly from the testicles, where sperm cells and their precursors keep the full genome throughout the animal’s life (lampreys throw away large chunks of the genome in most non-reproductive cells [Smith et al., 2009]). They probed these for Hox genes and sequenced the ones that tested positive. Plus they also got about two-thirds of the full genome together in fairly big pieces. Together, these data allowed them to get a better idea of the mess that is lamprey Hox cluster genomics.
They assembled four whole clusters, including their neighbouring genes, and a partial fifth cluster. A bunch of other genes sat on smaller sequence fragments containing only a couple of Hoxes, or a Hox and a non-Hox, but they were tentatively assigned to a total of eight clusters, eight being the number of different Hox4 genes in the data (no known vertebrate Hox cluster contains more than one Hox4 gene). The L. japonicum equivalents of the 31 publicly available Hox sequences from P. marinus spread out over six of these, which indicates that both species have at least six clusters. Seems like lampreys had another round of genome duplication after 2R? (Summary of L. japonicum Hox clusters from Mehta et al. below.)

But wait, that’s not the end of it.
First of all, although there are undoubtedly four complete Hox clusters in there L. japonicum, the relationships of these clusters to our four are terribly confused. Whether you look at the phylogenetic trees of individual genes, or the arrangement of non-Hox genes on either side of the cluster, only a big pile of what the fuck emerges. Phylogenies are problematic because the unusual composition of lamprey genes and proteins (Smith et al., 2013) could easily throw them off. All the complete lamprey clusters have a patchwork of neighbours that look like a mashup of more than one of our Hox clusters. Might it mean that lampreys’ proliferation of Hox clusters occurred independently of ours? Did we split before 2R after all?
Hox genes are not the only interesting things in a Hox cluster. In the long gaps between them, there are all sorts of little DNA switches that regulate their behaviour. Some of these are conserved across the jawed vertebrates. Mehta et al. aligned complete Hox clusters of humans, elephant sharks and lampreys to look for such sequences – called conserved non-coding elements or CNEs – in the lamprey.
They only found a few, but that’s enough for a bit more head-scratching. Most CNEs in, say, the human HoxA cluster are only found in one elephant shark cluster, and vice versa. Humans have a HoxA cluster, elephant sharks have a HoxA cluster, they’re clearly the same thing, pretty straightforward. Not so for lampreys. Homologues of individual CNEs in the complete lamprey clusters are spread out over all four human/elephant shark clusters. More evidence for independent duplications?
Mehta et al. are cautious – they point out that the silly mix of Hox cluster neighbours in lampreys could just be due to independent post-2R losses, which is plausible if the split between lamprey and jawed vertebrate lineages happened not too long after 2R. There’s also the fact that the weird lamprey sequences are phylogenetic minefields – however, that’s a double-edged sword, since the same caveat applies to analyses that support a post-2R divergence. Then, perhaps the same argument that goes for Hox cluster neighbours could also apply to CNEs. And, of course, this is just Hox clusters. Smith et al.‘s (2013) findings about overall genome structure don’t go away just because lamprey Hox clusters are weird.
So, in summary, thanks, lampreys. Fat lot of help you are! 😛
***
*Actually, two losses of two separate clusters in two different teleost lineages. Because Hox evolution wasn’t already complicated enough.
***
References
Kuraku S et al. (2009) Timing of genome duplications relative to the origin of the vertebrates: did cyclostomes diverge before or after? Molecular Biology and Evolution 26:47-59
Mehta TK et al. (2013) Evidence for at least six Hox clusters in the Japanese lamprey (Lethenteron japonicum). PNAS 110:16044-16049
Putnam NH et al. (2008) The amphioxus genome and the evolution of the chordate karyotype. Nature 453:1064-1071
Smith JJ et al. (2009) Programmed loss of millions of base pairs from a vertebrate genome. PNAS 106:11212-11217
Smith JJ et al. (2013) Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nature Genetics 45:415-421
Stadler PF et al. (2004) Evidence for independent Hox gene duplications in the hagfish lineage: a PCR-based gene inventory of Eptatretus stoutii. Molecular Phylogenetics and Evolution 32:686-694